
Application Note
High-throughput screening and selection of high lipid producers for biofuels
- Automated picking of up to 30,000 selected colonies per day
- User-defined parameters and flexible software for optimal colony picking
- Accurate, gentle colony picking at > 98% efficiency
- Variety of fluorescence filter sets for increased flexibility
Introduction
In recent years, there has been a markedly growing interest in alternative and renewable energy production products. One of the most prominent alternative energy resources is biodiesel, an energyrich portable fuel mainly composed of triacylglycerols (TAGs). Biofuels produced from crop seeds have recently came under major scrutiny due to the competition between foods vs. fuel. Biodiesel production from lipid producing microbial systems is receiving considerably more attention for generating sustainable and high-quality fuels. Oleaginous microorganisms (bacteria, algae and fungi) capable of accumulating significant quantities of stored TAGs are exploited as precursors for the production of lipidbased biofuels. Using synthetic biology and metabolic engineering approaches, researchers have engineered these microbes to produce next-generation biofuels and renewable energy products aimed to meet both environmental and market demands.
One of the major bottlenecks for biofuel production is robust identification of the high-value hit faster that is enabled by objective, functional readout of the end product in the primary screen. Although essential, manual colony picking followed by bicinchoninic acid assays, optical density measurements and gas chromatography assays can be time consuming, error-prone tasks. To enhance productivity and shorten timelines to find suitable candidates, automated and objective selection of colonies is required. Here we describe how QPix™ 400 Series provides a highly productive, reliable method to select desired lipidsecreting colonies based on quantitative measurement of lipid production with a fluorescent dye.
Culturing of lipid-accumulating bacteria
Rhodococcus opacus PD630 was selected as the model organism due to its high substrate tolerance, rapid growth, and ability to be cultivated to high densities. A strain of R.opacus (DSM 44193) was obtained from DSMZ culture collection, and cultured in Trypticase Soy Broth (TSB) medium at 28°C. For non-lipid accumulating negative control organism, a strain of Escherichia coli from ATCC (Migula 47076) was cultured in TSB medium at 37°C. Liquid cultures of both organisms were grown aerobically in a shaker incubator for 1-3 days. Strains were then cultured for 48-72 hours at 28°C on solid agar plates containing Trypticase soy agar (TSA) medium and 0.5 µg/mL of Nile red (AAT Bioquest, Inc.).
Objective colony screening using white light and fluorescence imaging
Nile red, a lipophilic fluorescent dye, detects lipid production directly in growing bacterial colonies on cultured agar plates and distinguishes between high lipid-accumulating R.opacus strain and non-accumulating negative control E.coli bacteria. The culture plates with colonies were imaged with QPix™ 420 system with white light and fluorescence using the FITC filter set (Ex 457nm/ Em 536nm) (Figure 1). Colonies containing high lipid-accumulating R.opacus strain displayed robust fluorescence under the UV fluorescence channel on the QPix 420 system, while negative control colonies exhibited no significant fluorescence (Figure 2). Colony detection using both white light and fluorescence imaging on the QPix System enables objective identification and selection of colonies that exhibit fluorescence based on lipid accumulation.
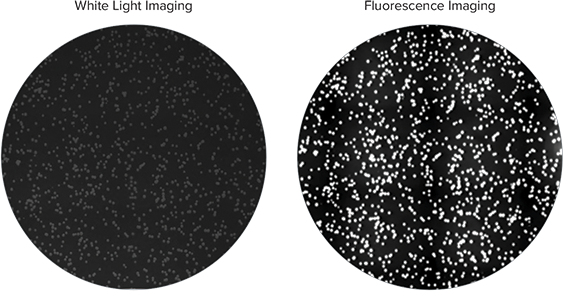
Figure 1. Colonies of R.opacus grown in media containing Nile red were imaged on QPix 420 system under white light (left) and fluorescence imaging (right). Colonies accumulating high amount of lipids can be detected based on fluorescence intensity.
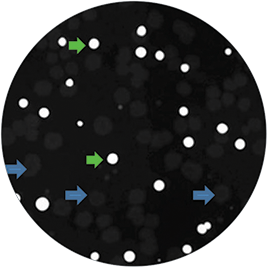
Figure 2. Non-lipid accumulating E.coli (negative control) were co-cultured with high lipid accumulating R.opacus on media containing Nile red and imaged on QPix 420 system with fluorescence imaging module. Green arrows depict colonies accumulating high amounts of lipid as observed by bright fluorescence; blue arrows indicate negative control colonies that do not exhibit any fluorescence.
Optimized colony selection using tunable software parameters
The acquired images were analyzed using the QPix Software. Parameters such as size, diameter and compactness were tuned for optimal colony selection. A mean fluorescence intensity threshold of greater than 50,000 was set to select (i.e. gate) colonies demonstrating high fluorescence as a result of high lipid accumulation detected by staining with Nile red (Figure 3).
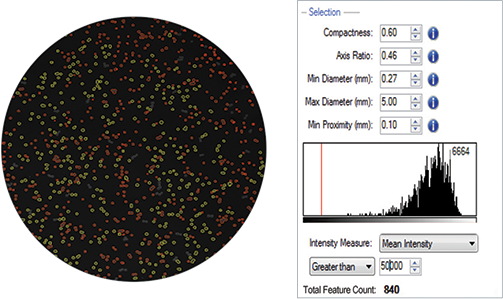
Figure 3. Fluorescence-based quantification that is reflective of lipid accumulation would enable objective colony selection on QPix 400 Series with user defined colony selection criteria (red).
Both fluorescent and non-fluorescent colonies indicating high and no lipid accumulation, respectively, were picked using the QPix System’s fully pneumatic 96-pin picking head. Lipid accumulation levels were further confirmed postpicking. Following an overnight culture in liquid media, cultures were stained with a lipophilic bright green fluorescent dye, BODIPY 505/515 (Life Technologies) at a concentration of 0.5 µg/mL and fluorescence measurements were recorded on SpectraMax® M5 MultiMode Microplate Reader. A high degree of fluorescence was confirmed in high lipid accumulating colonies that were originally selected and picked using the QPix 420 System, while background levels of fluorescence were exhibited by the negative control E.coli colonies as depicted in red (Figure 4A). Additionally, images of BODIPY 505/515-stained high lipid producing R.opacus were acquired using ImageXpress® Micro XLS Widefield High Content Screening System at 40x magnification (Figure 4B). Bright green fluorescent staining with lipophilic dye is indicative of high lipid accumulation in R.opacus.
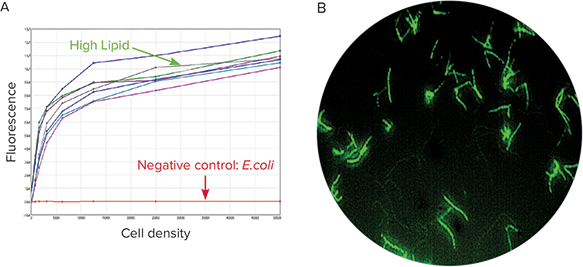
Figure 4. (A) Fluorescence reading of high lipid producing colonies and negative control group stained with BODIPY 505/515 lipophilic fluorescent dye on the SpectraMax M5 Multi-Mode Microplate Reader. High lipid producing colonies exhibited high fluorescence (RFU) values and in contrast the negative colonies exhibited a flat-line signal as depicted in red. (B) Fluorescent image of high lipidyielding R.opacus demonstrated bright green staining with lipophilic BODIPY 505/515 dye as acquired using the ImageXpress Micro XLS Widefield High Content Screening System at 40x magnification.
These data demonstrate the ability to differentially select and pick colonies based on fluorescence exhibited by the high lipid producing colonies. The ability to examine large numbers of colonies for applications such as library screening enables efficient and reliable identification of high-value vs. low-value strains. Additionally, with quantitative fluorescence imaging and objective software analysis provided by the QPix 400 Series, secondary screening assays could potentially be eliminated from the workflow. This would save sample processing time and reduce costs while improving the workflow efficiency to isolate high-value colonies.
Summary
The QPix 400 Series of automated microbial colony pickers provides a highly productive and reliable alternative to the conventional manual picking approaches. Implementing the QPix 400 Series in experimental workflows may shorten the overall processing time to obtain the colonies of interest. With the aid of fluorescence detection capabilities of QPix 400 Series, high lipid producing bacterial colonies can be accurately detected and selected. Flexible and easy-to-use software allows users to customize and define optimal colony selection criteria for automated picking experiments. This work describes a time-saving approach to isolate high-value colonies in the study of metabolic engineering or protein evolution for biofuel production.