
Application Note
Increase Cell Painting assay throughput using an automated workstation
- Reduce hands-on time for the Cell Painting assay by over 80%
- Flexible, automated process can be readily modified for a wide variety of applications
- Reduce human errors and enhance throughput in the Cell Painting assay
Abstract
Michael Hayes, PhD | Beckman Coulter Life Sciences
Angeline Lim, PhD | Molecular Devices
Cell painting has become a popular screening method within the drug discovery community. This phenotypic screening platform leverages advances in high-content microscopy, high-throughput screening, liquid handling, and computation to generate massive amounts of data. Cell painting assays are often used in combination with genetic perturbations and/or drug-like small molecule libraries in order to identify new drugs or drug targets capable of inducing a phenotype of interest. Cell painting workflows can be time- and labor-intensive, taking several days and requiring screening of many assay plates. The use of automated liquid handlers, like the Biomek i7 Hybrid automated workstation could help to streamline these processes, saving valuable user time and increasing assay throughput. Here we developed an automated workflow to prepare cell painting assay plates. After staining the cells, the Molecular Devices ImageXpress® Confocal HT.ai High-Content Imaging System was used to acquire high-quality images for downstream data analysis. Using a Biomek i7 hybrid automated workstation, cells were paraformaldehyde fixed, permeabilized with Triton X-100, and treated with a combination of six spectrally compatible fluorescent stains. We also performed a colchicine time course experiment as a case study to simulate a small molecule screening setup. Together, the data presented here highlights how the use of an automated workflow combining Biomek liquid handling with ImageXpress imaging can be used for morphological profiling, with the added benefits of reduced hands-on time and user handling errors with increased assay throughput.
Introduction
Recent advances in cell biology have allowed the development of increasingly complex models of human disease. These advances include new cellular models, like patient-derived induced pluripotent stem cells, new culture methods, like three-dimensional organoids, and new, precise genetic editing methods, like CRISPR/Cas9. Many workflows involving these recent cellular models use fluorescent imaging as a final readout, which has led to the development of more complex, information-rich experimental setups. One workflow that is gaining traction within the drug discovery community is cell painting, which is a cellular morphology profiling method that uses common, commercially available fluorescent dyes in a multiplexed manner.1This phenotypic screening platform leverages advances in high-content microscopy, highthroughput screening, and liquid handling to generate massive amounts of data. Cell painting has been used for analyzing a variety of cellular manipulations, like genetic and small molecule screening. In a genetic screen, each well contains reagents to manipulate a gene of interest, often using siRNA or CRISPR/Cas9. In a small molecule screen, thousands to millions of compounds are tested to identify drug-like lead compounds.
Following the execution of a cell painting screen, imagebased profiling can be used to cluster genes/compounds based on mechanism of action.2 Further, there is a growing amount of interest in using artificial intelligence and machine learning to identify biologically active compounds in physiologically relevant model systems based on these high-content images.3 Traditional screening campaigns use a single readout/measure to identify the effect of each gene/compound on the phenotype of interest. Alternatively, cell painting has the advantage of creating a morphological cell profile for each condition tested. Post hoc analyses can then be used to cluster known, control gene/drug conditions with experimental treatments based on the cell painting profiles observed.4 (This is an area of research that will see increased utility in the drug discovery community as the computational methodologies are refined with more image-based datasets.)
The general workflow for a prototypical cell painting assay is outlined below (Figure 1). First, cells are plated into optical bottom, microtiter plates. In general, adherent cells are preferred, as the imaging portion of the workflow becomes more straightforward. Cells are then treated with the experimental conditions of interest, either via transfection (genetic screen) or compound addition (small molecule screening) and incubated until the phenotype of interest can be observed. The cells are then fixed (paraformaldehyde), permeabilized (Triton X-100), and stained (if live stains are used, this is done before fixation). Plates are then imaged using an automated, fluorescent confocal microscope, like the Molecular Devices ImageXpress Confocal HT.ai High-Content Imaging System. The images are then analyzed within image analysis software such as IN Carta® Image Analysis Software or the open source CellProfiler software package, and the measurements are fed into a data mining pipeline that can generate analyze cell morphology profiles, such as the web-based StratoMineR (Core Life Analytics).5

Figure 1. Cell Painting Workflow Simplified protocol for automated cell painting liquid handling, image acquisition, and data analysis. Cells were plated, drug treated, fixed, and stained using a Biomek i7 hybrid automated workstation. Images were acquired using the Molecular Devices ImageXpress Confocal HT.ai.
This workflow takes several days and requires several touchpoints by the user. Performing all these steps with manual pipetting is possible when testing only a small number of conditions or working in 96-well plates, but when higher density (384-well or more) or increased throughput is required, performance of these steps by hand becomes impractical. The Biomek i7 hybrid automated workstation is an automated liquid handler that is capable of efficiently performing the complex liquid handling steps of cell painting workflows (Figure 2). This minimizes the number of required user interactions and increases walkaway time, freeing the operator to attend to other laboratory tasks. The Multichannel pod can be equipped with 96- or 384-well heads that can accurately pipette 1 to 1200 μL and 0.5 to 60 μL, respectively. Additionally, the 8-channel Span-8 pod is accurate from 0.5 to >1000 μL. This workstation supports 45 deck positions and can be directly fitted with devices, like orbital shakers, heating/cooling Peltiers, and tipwashers for plate and sample processing (Figure 2).

Figure 2. Beckman Coulter Life Sciences Biomek i7 Hybrid Automated Workstation and the Molecular Devices ImageXpress Confocal HT.ai High-Content Imaging System.
Further, depending on user needs, the Biomek i7 Hybrid workstation supports integration with other automated plate handling instruments, such as tissue culture incubators, barcode readers, plate washers, bulk reagent dispensers, multimode plate readers, centrifuges, automated fluorescent microscopes, and more. Another important feature is the optional HEPA filter, which creates a more sterile environment, an important factor when handling mammalian cell cultures. Thus, a Biomek system integrated with an automated cell incubator (e.g., Liconic, Cytomat, Thermo Fisher) and a high-content imager can perform the entire cell painting workflow from cell plating to image acquisition, only requiring deck setup from the user. The ImageXpress Confocal HT.ai High- Content Imaging System is a scalable high-throughput and high-content screening solution equipped with a 7-laser light source. Use of high-intensity lasers allows for shorter exposure times without compromising image quality. This translates into significant improvement in acquisition speed compared to systems with LED light sources. In addition, the ImageXpress Confocal HT.ai can image up to 8 channels, which makes it ideal for highly multiplexed assays such as cell painting.
Here, we demonstrate an automated workflow with HEK293 cells, from cell plating to fixation and staining on a Biomek i7 hybrid workstation, followed by image acquisition using an ImageXpress Confocal HT.ai High-Content Imaging System. The automated workflow can reduce hands-on time and the possibility of sample-handling errors by the user, while also increasing throughput.
Methods
Cell culture
Lenti-X 293T cells (Takara) were maintained at 5% CO2 and 37°C in growth medium, which was composed of DMEM supplemented with 10% FBS and 1% 100X antibiotic/ antimycotic (Gibco). For cell harvest, cultures were washed with DPBS, dissociated with trypsin, pelleted at 300 x g in an Allegra X-14R centrifuge (Beckman Coulter Life Sciences), and resuspended in 1:1 Opti-MEM I to growth medium. Cells were counted using a hemocytometer and further diluted in Opti-MEM I to achieve the desired cell density of 100 cells/μL. For 96-well plates, cells were plated at 10,000 cells per well (100 μL total volume) using a Biomek i7 hybrid workstation equipped with HEPA filtration unit. In order to simulate small molecule drug treatment, colchicine was diluted to 1 μM in Opti-MEM I and 50 μL was added to each well of the 96-well cell painting assay plate at the indicated time point using the Biomek i7 hybrid workstation. Opti-MEM I without colchicine served as control.
Automated cell plate preparation
In general, the experiments performed here followed previously described protocols.1 For the automated liquid handling method, all aspiration and wash steps were performed using a 96-well Multichannel pod equipped with a 1200 μL head, and all reagent addition steps were performed with the Span-8 pod. The deck layout for the automated method is presented in Figure 3. All automated liquid handling steps were performed using a pipetting template that aspirated and dispensed at 5 μL/sec at a position >70% away from the center of the plate well. This ensured that cell monolayers on the bottom of the wells were undisturbed during the cell painting procedure. Paraformaldehyde solution was made fresh the day of the assay by dilution of a 16% stock solution to 6.8% PFA in ultra-pure water.
First, 110 μL of well contents was aspirated to leave a final volume of 40 μL in the 96-well plates, and wells were washed twice with 110 μL of DPBS. Next, 60 μL of MitoTracker Deep Red was added to each well, the plate was incubated in the dark at ambient temperature for 20–30 minutes, and the cells were washed again with 110 μL of DPBS. Next, the cells were fixed by the addition of 60 μL of 6.8% PFA, to achieve a final concentration of 4% PFA. Plates were incubated in the dark at ambient temperature for 20 min, PFA was removed, and cells were washed twice with 110 μL of DPBS, leaving a final volume of 150 μL of DPBS in each well. Plates were then sealed and stored in the dark at 4°C until the day of the cell painting assay, when cells were stained and imaged.
Cell painting assay
Triton X-100 permeation solution was made by diluting Triton X-100 to 0.1% in DBPS supplemented with 0.1 mg/mL BSA. Hoechst, concanavalin A, SYTO 14, WGA, and phalloidin fluorescent dye stock solutions were prepared and combined into a single staining master mix as previously described in Bray et al. (1). Cells were washed with 110 μL of DPBS then permeabilized by the addition of 120 μL of Triton solution for 10–20 minutes in the dark at ambient temperature. Triton was then removed, and cells were washed twice with 110 μL of DPBS. Cells were stained by the addition of 40 μL of staining reagent for 30 minutes in the dark at ambient temperature. Finally, stain was removed, cells were washed three times with 110 μL of DPBS, and plates were imaged using the ImageXpress Micro HT.ai High-Content Imaging System.
Image acquisition and data analysis
Images were analyzed using IN Carta Image Analysis Software. The image segmentation protocol was set up as follows: Nuclei was assigned as the primary target using the custom segmentation pre-trained model Nuclei.a.h5. Objects touching edges were excluded. Cells were segmented using the YFP channel (SYTO14) with the Robust option. Three additional organelle classes were segmented with the indicated option: Mitochondria (networks), Actin (fibers) and endoplasmic reticulum (networks). Measurements from each cellular compartment were extracted. Measurement such as shape, size, intensity, texture, colocalization and spatial relation to neighboring objects were selected as analysis output. In total, 487 features were extracted per cell.
For the data analysis, cell-level data was exported in CSV format and then uploaded into HC StratoMineR (CoreLife Analytics) along with a text file that contains metadata with compound information. Plate map was defined within the StratoMineR interface. Using the Quality Control tab, outlier wells can be removed from analysis (wells with less than 50 cells were removed). Data transformation was performed as recommended followed by feature scaling. Principal component analysis (PCA) was used for data reduction. The resulting 7 components were used to calculate the distance score, which is defined as measure of phenotypic effect of the treatment on the cells in that wells.6
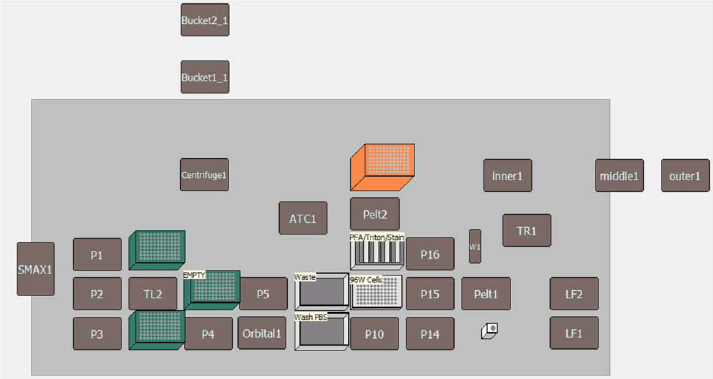
Figure 3. Deck Layout for Automated Cell Painting Assay The method automated using a Biomek i7 hybrid workstation used the following components: three tip loading ALPs, five 1X1 ALPs, Multichannel pod, Span-8 pod, 2 boxes 230 μL pipette tips (plus 1 empty box), and 1 box 1070 μL pipette tips..
Results and discussion
Cell painting has been gaining popularity within the screening community, as it can provide information-rich results following a variety of cellular manipulations. The plate preparation workflow for cell painting assays involves labor intensive liquid handling steps that can become impractical to perform manually, especially as the number of plates being screened increases (1). As such, we developed a walkaway plate processing method using a Biomek i7 hybrid workstation using previously described best practices (1). Prior to painting the plates, cells were fixed and permeabilized using 4% PFA and 0.1% Triton X-100, respectively. Here we compare the liquid handling of our newly developed automated method with cells painted by hand.
The first parameter that was evaluated with the automated workflow was the time savings versus the manual workflow. A single 96-well plate was painted manually using a 12-well multichannel pipette and with the Biomek i7 Hybrid workstation. For a single plate, both workflows took approximately 2 hours to complete (120 manual vs 115 min Biomek), and 60 minutes of the total time was incubations at ambient temperature in the dark. For the Biomek method, the only hands-on time required was 5 to 10 minutes at the beginning of the method for deck setup. Alternatively, the manual method took 60 minutes of active pipetting by hand (Table 1). Further, the automated method was easily adaptable to 384-well format. The 384-well method took approximately 120 min, but again deck setup was the only hands-on step required (5–10 min). The 384-well workflow was not performed by hand, as this was deemed impractical because this plate format could require up to 3 to 4 hours of manual pipetting.
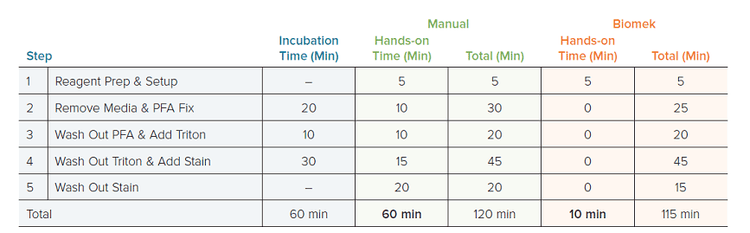
Table 1. Timing of Manual vs Automated Cell Painting
In order to qualitatively evaluate this newly developed automated method, we stained HEK293 cells with several commercially available fluorescent stains (Figure 4). These results largely exhibited the staining pattern that was expected. The endoplasmic reticula, Golgi, mitochondria, nuclei, and RNA (nucleoli) exhibited staining patterns consistent with previously reported cell painting results (Figure 4) (1). Interestingly, f-actin staining with phalloidin resulted in robust fluorescent labeling but distinct actin filaments were not discernable as in previous cell painting results. This could be due to the combination of the HEK cell line and the concentration of phalloidin stain used here. This highlights that prior to performing large-scale screens, preliminary experiments may be required to identify optimal stain concentrations for the cell line of interest. More importantly the results from this experiment indicated that the automated cell painting and image acquisition workflow worked. Cells were clearly present on the bottom of each well, indicating that the automated liquid handling was gentle enough to keep cells adhered to the plate while also successfully fixing the cells. Further, several of the dyes (phalloidin, concanavalin) are normally impermeable to cells, so the staining of intracellular compartments indicates that adequate cell permeability was achieved using 0.1% Triton. Finally, the robust fluorescent signal observed in each channel shows that the staining step was also successful.
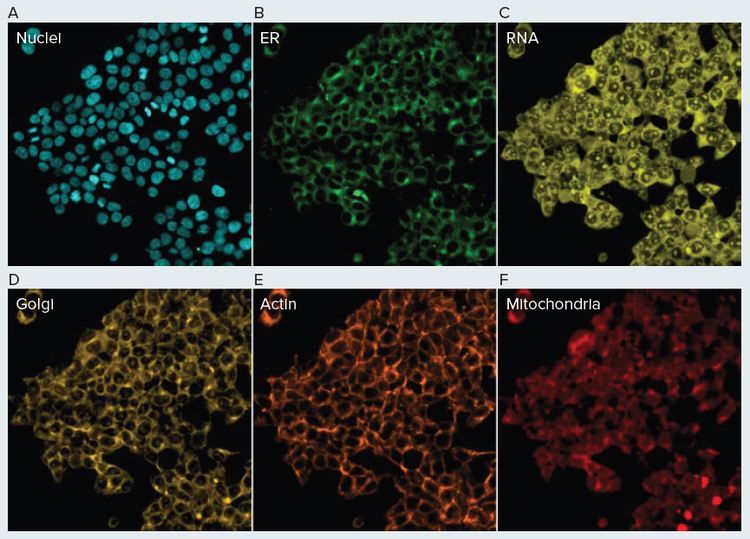
Figure 4. Automated Cell Painting Assay HEK293 cells were painted using a Biomek automated method and imaged using the Molecular Devices ImageXpress Confocal HT.ai High-Content Imaging System. Endoplasmic reticula were stained with concanavalin A (A), Golgi/plasma membrane with wheat germ agglutinin (B), mitochondria with MitoTracker (C), nuclei with Hoechst 33342 (D), actin with phalloidin (E), and RNA with SYTO Green (F).
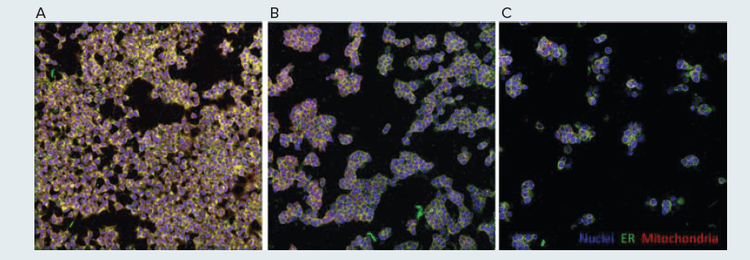
Figure 5. Simulated Cell Painting Screening Controls HEK293 cells were treated with vehicle control (A) or 300 nM colchicine for 4 hours (B) or 24 hours (C). Images were acquired on the Molecular Devices ImageXpress Confocal HT.ai High-Content Imaging System.
As cell painting is often used as a screening technique, each well will often contain a different cellular manipulation, such as a different drug-like small molecule. In order to simulate this assay setup, colchicine was selected as a control compound. Colchicine binds and inhibits tubulin, which is required for mitosis and subsequent normal cell division. Due to this mechanism of action, treatment with this drug was predicted to induce a dramatic change in the morphology of HEK293 cells. Using the Biomek, cells were plated and treated with 300 nM of colchicine for either 24 hr or 4 hr, and untreated cells were used as control. Following treatment, cells were painted and imaged using the Biomek i7 hybrid workstation and ImageXpress Confocal HT.ai High- Content Imaging System, respectively. The difference in cellular phenotype observed between control and colchicine-treated cells was dramatic (Figure 5A, 5C). Cells that were treated with colchicine appeared larger and rounder, indicative of cell cycle arrest during the metaphase of mitosis. Further, there were fewer cells per well following drug treatment, likely due to the inability of cells to properly divide.
Another important part of cell painting workflow is the downstream image and data analyses. The images were first analyzed using In Carta Image Analysis Software. IN Carta features a deep learning-based segmentation tool (SINAP) that enables robust feature identification. For the analysis, the nuclei model in SINAP improved nuclei detection and was able to accurately split touching nuclei. The SYTO14 stain was used to define the cytoplasm and the ER, mitochondria and actin structures were also identified and analyzed. Overall, 487 measurements representing intensity, texture, shape, spatial relationship between organelles, and colocalization numbers were derived from each cell.
For each condition shown in Figure 5, cell level data from 24 replicate wells were fed into the HC StratoMineR (Core Life Analytics) cloud-based tool. Principal component analysis was used to reduce the 487 measurements in 7 principal components. Three distinct clusters were observed, and each cluster was representative of a single colchicine treatment condition (Figure 6). This highlights the utility of automated liquid handling and image acquisition in cell painting workflows, as each replicate was accurately clustered with identically treated wells. This also highlights the utility of quality control compounds, as colchicine induced robust time-dependent phenotypic differences that could be distinguished with analysis using IN Carta and HC StratoMineR.
The results presented above were for processing a single plate, but many workflows will require processing of multiple plates in order to screen larger libraries. This is another area where the use of automated liquid handlers can excel. The Biomek workstation can be fitted with various integrations to help perform automated cell handling and imaging, like automated incubators (Cytomat) and high-throughput fluorescent microscopes (Molecular Devices). In our hands the use of slow, gentle pipetting techniques achievable with the Biomek were preferred to other methods, but some users may prefer to perform the cell washing steps using plate washers (BioTek 405) or bulk reagent dispensers (MultiFlo/ Multidrop). Fortunately, the Biomek iSeries can also be integrated with these instruments, allowing for more advanced, higher throughput workflows than the one described here. Additionally, fluorescent imaging of cell painting plates can take a long time (1-2 hr per plate), so some users may not want to assemble multiple plates at one time. Beckman Coulter Life Sciences also offers SAMI EX scheduling software. This software tool can be used to schedule advanced timing and plate handling operations to interleave method components, like staining and plate reading, to ensure that all plates are treated to approximately the same time course during fixation/ permeation/staining. Together these hardware and software options could provide high-throughput, walkaway cell painting workflows beyond what is demonstrated here.
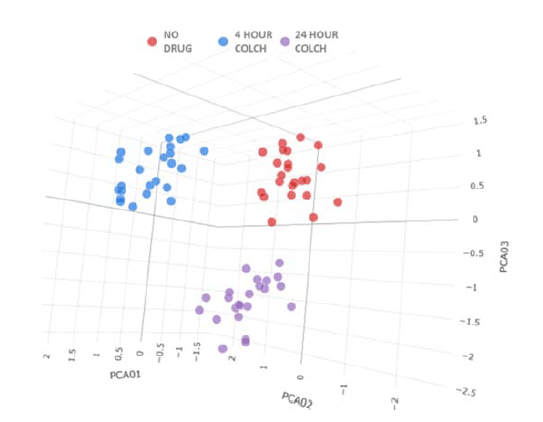
Figure 6. Data PCA Plot For each condition, 24 replicate wells were evaluated in HC StratoMineR. Wells were separated into three distinct clusters, each representative of one treatment condition.
Summary
As the drug discovery community’s ability to generate highly complex cell models of disease has advanced, so too has the interest in generating highly complex assay readouts. One method that has garnered attention is the cell painting technique, in which a few simple, commercially available cell stains are multiplexed to generate complex phenotypic data(1). By combining this phenotypic method with genetic (siRNA/CRISPR) and small molecule screening efforts, vast amounts of data can be generated from a single experiment. Following the execution of a cell painting screen, image-based profiling can be used to cluster genes and/or compounds based on phenotype to predict a putative mechanism of action(2). Unfortunately, two bottlenecks remain in the cell painting workflow: preparation of assay plates and automated image acquisition.
Preparing assay plates manually is a time- and laborintensive process that can take up to several days, depending on the time course of drug treatment/ transfection. Further cell fixation/permeation/staining can require up to 1 hr of hands-on time per 96-well plate. To address these obstacles, we sought to automate a cell painting workflow from cell plating and treatment to fluorescent staining to image acquisition. Here we highlight the utility of this workflow and the capabilities of the Biomek i7 hybrid workstation and ImageXpress Confocal HT.ai High-Content Imaging System. Initial experiments showed that plates were readily cultured, fixed, stained, and imaged using the automated method. Subsequent work showed that small molecule (colchicine) screening workflows could be automated, and meaningful cell painting data can be obtained using this method. Together, the use of Biomek automation and ImageXpress imaging provides value for cell painting applications by reducing hands-on time and increasing assay throughput.
References
- Bray, M. A., Singh, S., Han, H., Davis, C. T., Borgeson, B., Hartland, C., Kost-Alimova, M., Gustafsdottir, S. M., Gibson, C. C., & Carpenter, A. E. (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols, 11(9), 1757–1774. https://doi.org/10.1038/nprot.2016.105.
- Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D., & Carpenter, A. E. (2021). Image-based profiling for drug discovery: due for a machinelearning upgrade?. Nature Reviews Drug Discovery, 20(2), 145–159. https://doi.org/10.1038/s41573-020-00117-w.
- Simm, J., Klambauer, G., Arany, A., Steijaert, M., Wegner, J. K., Gustin, E., Chupakhin, V., Chong, Y. T., Vialard, J., Buijnsters, P., Velter, I., Vapirev, A., Singh, S., Carpenter, A. E., Wuyts, R., Hochreiter, S., Moreau, Y., & Ceulemans, H. (2018). Repurposing High-Throughput Image Assays Enables Biological Activity Prediction for Drug Discovery. Cell Chemical Biology, 25(5), 611–618.e3. https://doi.org/10.1016/j.chembiol.2018.01.015.
- Gustafsdottir, S. M., Ljosa, V., Sokolnicki, K. L., Anthony Wilson, J., Walpita, D., Kemp, M. M., Petri Seiler, K., Carrel, H. A., Golub, T. R., Schreiber, S. L., Clemons, P. A., Carpenter, A. E., & Shamji, A. F. (2013). Multiplex cytological profiling assay to measure diverse cellular states. PloS One, 8(12), e80999. https://doi.org/10.1371/journal.pone.0080999.
- Kamentsky, L., Jones, T. R., Fraser, A., Bray, M. A., Logan, D. J., Madden, K. L., Ljosa, V., Rueden, C., Eliceiri, K. W., & Carpenter, A. E. (2011). Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics (Oxford, England), 27(8), 1179–1180. https://doi.org/10.1093/bioinformatics/btr095
- Omta WA, van Heesbeen RG, Pagliero RJ, van der Velden LM, Lelieveld D, Nellen M, Kramer M, Yeong M, Saeidi AM, Medema RH, Spruit M, Brinkkemper S, Klumperman J, Egan DA. HC StratoMineR: A Web-Based Tool for the Rapid Analysis of High-Content Datasets.Assay Drug Dev Techno l. 2016 Oct;14(8):439-452. doi: 10.1089/ adt.2016.726. Epub 2016 Sep 16. PMID: 27636821.
Materials
Table 2. Instruments used
Table 3. Reagents used
Table 4. Consumables used per run